Introduction
ATAC-seq(Assay for Transposase Accessible Chromatin with high throughput sequencing) This technology can capture and sequence the open regions of chromatin structure through transposase with high activity after transformation, and only a few cells are needed to obtain real-time genome-wide activity regulation sequence information. This technology uses transposase to cleave the open nuclear chromatin region in a specific space-time, and then obtains all the active transcriptional regulatory sequences in the genome in that specific space-time.
Eukaryotic DNA is not naked, but is packaged into nucleosomes to form Beaded structures and further folded and packaged. For the transcription of genes, it is necessary to unravel this high-level structure, so that DNA can become a bare state with which various transcription machines can bind, that is, to form open chromatin regions. ATAC SEQ, as a new technology for studying open chromatin, uses Tn5 transposase to enter and cleave naked DNA, and at the same time connects specific sequencing adapters. Because cutting and jointing are completed in one step, this technology can greatly reduce the required cell starting amount.
Technical advantages
1. low initial amount of cells required
- Wide range of applications, suitable for most species and cell types
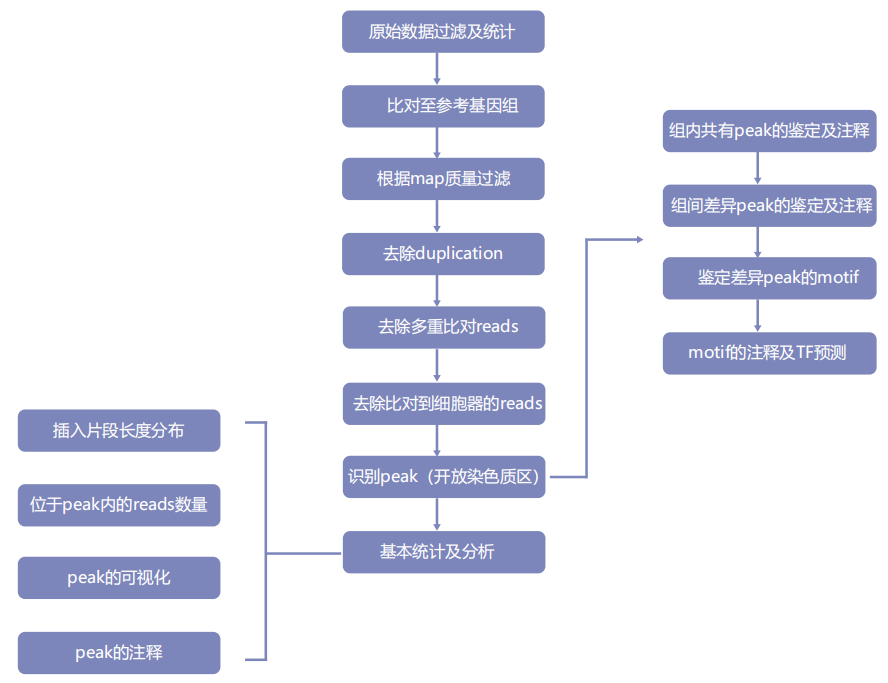
Experimental protocol

Information analysis
Technical parameter
| Sample requirements | Project cycle |
| The cells in good growth condition were trypsinized and counted to ensure that the cell viability was above 85%; Resuspend the cells in the cryopreservation solution, and add 80000-100000 cells (1ml cryopreservation solution) to each cryopreservation tube. The number of cells should not be too many or too few, and try to be in this range, otherwise the effect will be affected); Put the cryovial into the programmed cooling box and directly put it into the -80 ℃ refrigerator. Notes: 1. it is recommended to use the programmed cooling box. First, it can reduce the operation steps. Second, the effect is better than the traditional isopropanol, which can be purchased from the reagent dealer. 2. basic formula of cryopreservation solution: 50% fbs+40% complete medium +10% dmso. (the concentration of DMSO should not be higher than this, otherwise it will damage cells and lead to increased mortality) 3. ensure sufficient dry ice for transportation, and put the cells in the middle to avoid cell death caused by warming. | The operation cycle of the standard process below 15 samples is about 50 working days. When the number of samples is large, the project cycle needs to be determined with the technical support personnel. |
Case sharing
Case:
ATAC SEQ analysis revealed a general decrease in chromatin accessibility in patients with age-related macular degeneration
Wang J, Zibetti C, Shang P, et al. ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration[J]. Nature Communications, 2018, 9(1).
Background:
Age related macular degeneration (AMD) is the main factor leading to the loss of vision in the elderly, most of which occur over 45 years old, and its prevalence increases with age. At present, the epigenetic changes in the course of AMD are not clear. In this paper, ATAC SEQ was used to study the chromatin accessibility of retina and retinal pigment epithelial cells (RPE) in patients with AMD.
Methods:
The open chromatin regions of retina and RPE in AMD and normal tissues were identified by ATAC SEQ. In addition, RNA SEQ technology was used to analyze the relationship between open chromatin regions and gene expression.
Result:
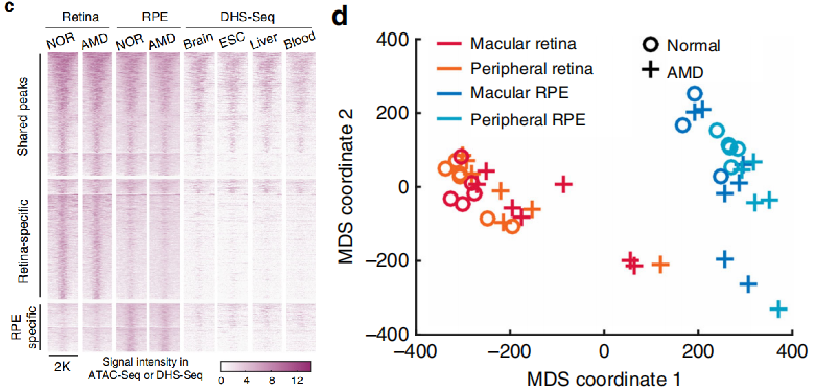
The identified peaks (open chromatin regions) were classified to obtain retina specific, RPE specific and shared peaks. Compared with different tissues (DNase SEQ data), it was found that most of the shared peaks were tissue-specific, while most of the retina and RPE specific peaks were tissue-specific (left panel). Using peaks to classify the samples, we found that retina and RPE were obviously separated, and normal tissues and AMD also tended to be separated (figure below).
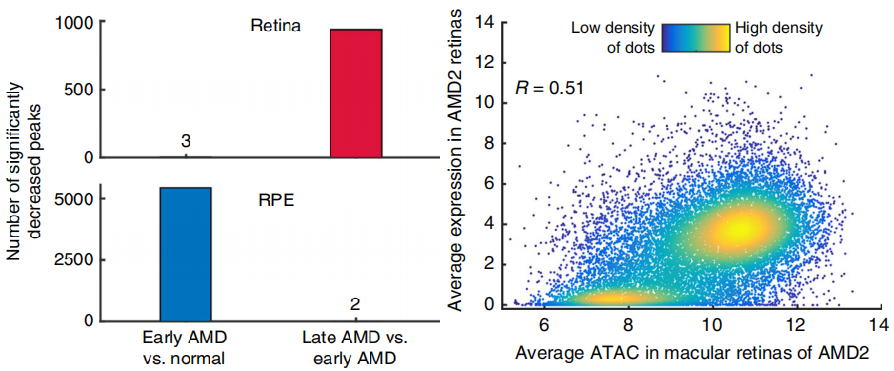
The differential open chromatin regions (AMD) of retina and RPE in early AMD vs normal tissues and in late amd vs early AMD were identified respectively, indicating that retina chromatin accessibility decreased mainly from early AMD to late AMD, while RPE chromatin accessibility decreased mainly from normal tissues to early AMD (lower left panel). Combined with transcriptome analysis, ATAC SEQ peaks intensity and gene expression showed a strong correlation, with a correlation coefficient of 0.51 (lower right panel).