Introduction
Target region capture bisulfite methylation sequencing is to specifically capture the specific region of the genome of interest by designing the corresponding probe sequence. The captured fragment is further treated with bisulfite and the methylation level of the target region is detected by high-throughput sequencing. This technology is the first choice for large sample size and fixed region methylation detection, and can be widely applied to the identification of methylation specific regions of clinical samples and the screening and validation of disease-related methylation bio marker.
Technical advantages
1.Multiple coverage sites
Acquire more CpG sites than methylation chip
2. wide detection range
In addition to covering known DMR regions, new DMR regions can be found
3. high accuracy
Accurate to single base resolution and highly consistent with published genome-wide methylation sequencing data
4. High accuracy
The reproducibility of target region enrichment, sequencing depth distribution and coverage is high; Agilent sureselectxt human methyl SEQ capture probe is preferred, and the target regions include:
① The target region was 84m, and the CPG site of 3.7m was detected;
② It mainly targets CpG island, gencode promoter and known regulatory regions;
③ Covering known cancer specific DMR regions and tissue-specific DMR regions;
④ It contains the "shores" and "shells" regions 4KB upstream and downstream of CpG island;
⑤ Contains deoxyribonuclease I (DNase I) high-sensitivity sites. The whole genome coding gene promoter chip (2k upstream and 500bp downstream of TSS, nimblegene chip) can also be used.
Experimental strategy

Information analysis
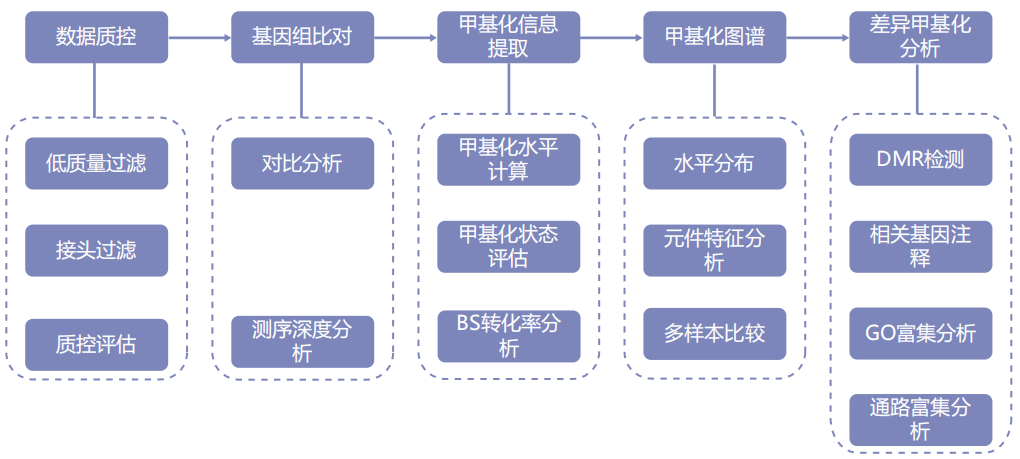
Technical parameter
| Sample requirements | Project cycle |
| Total amount: different samples have different requirements, human samples ≥ 2 μ g; Purity: od260/280 = 1.8~2.0; Integrity: there is no obvious degradation of DNA, and gel electrophoresis detection gel map shall be provided.Standard chips are available: ① whole genome coding gene promoter chip (2k upstream and 500bp downstream of TSS, nimblegene chip) ② Agilent sureselectxt human methyl SEQ chip | The operation cycle of the standard process below 15 samples is about 50 working days. When the number of samples is large, the project cycle needs to be determined with the technical support personnel. |
Case analysis
案例:
DNA methylation of hepatocellular carcinoma based on bisulfite sequencing captured by liquid phase hybridization
Gao, F., et al. (2015).Global analysis of DNA methylation in hepatocellular carcinoma by a liquid hybridization capture-based bisulfite sequencing aproach. Clin Epigenetics 7: 86.
Background:
The sequencing cost of whole genome bisulfite methylation sequencing is high, and the modification research of DNA methylation often only focuses on the region near the gene. Selective capture and bisulfite sequencing of the region of the genome can effectively reduce the sequencing cost, and it is expected to be applied to the research field of cancer epigenomics.
Methods:
Based on bisulfite sequencing technology of target region liquid-phase capture, the promoter DNA methylation of hepatocellular carcinoma was studied.
Conclusion:
A technology based on the combination of whole genome promoter region liquid-phase hybridization capture and bisulfite sequencing was established, and the DNA methylation level of the whole genome promoter region of eight hepatocellular carcinoma patients and adjacent tissues was studied by using this technology, involving a total of 1.86mb CpG sites. At the same time, combined with gene expression data and later large-scale sample validation, seven differentially methylated genes related to the occurrence and development of liver cancer were successfully identified.
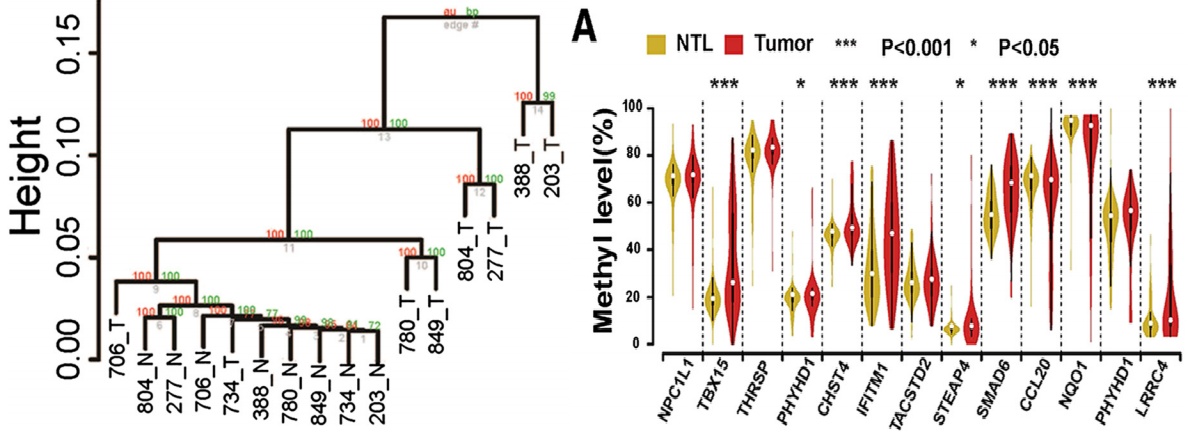
8 cluster analysis of DNA methylation and differentially methylated candidate genes in cancer and adjacent tissues of hepatocellular carcinoma patients


