Introduction
Chromatin co immunoprecipitation sequencing (chip SEQ) is a chip SEQ technology combining chip and high-throughput sequencing technology, which can efficiently and accurately screen and identify the DNA binding sites of specific proteins in the whole genome. The differential modification of different histones is closely related to gene transcription, spatial configuration and conformation of chromatin. The specific antibody of a modified specific histone is used to enrich the DNA fragment bound to it, and the purification and library construction are carried out, followed by high-throughput sequencing. By accurately comparing the obtained data with the reference genome, researchers can obtain the relationship between a specific histone of a modified type and the genomic DNA sequence in the whole genome, and can also make differential comparison of multiple samples.
Technical advantages
1. wide screening range
Genome wide screening for the relationship between specific histone modifications and genomic DNA sequences
2. trace samples
Only 5ng or more of immunoprecipitated DNA was needed to start sequencing analysis
3. flexible scheme
Select different technical parameters of histone modification specific antibodies according to different project requirements
Experimental protocol
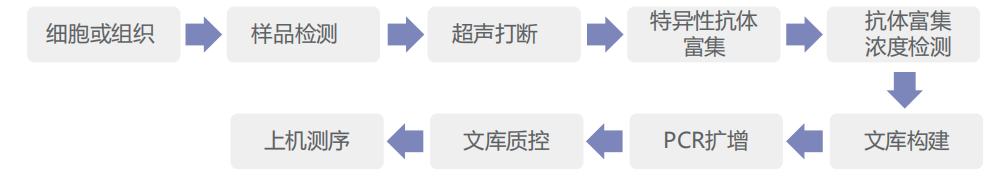
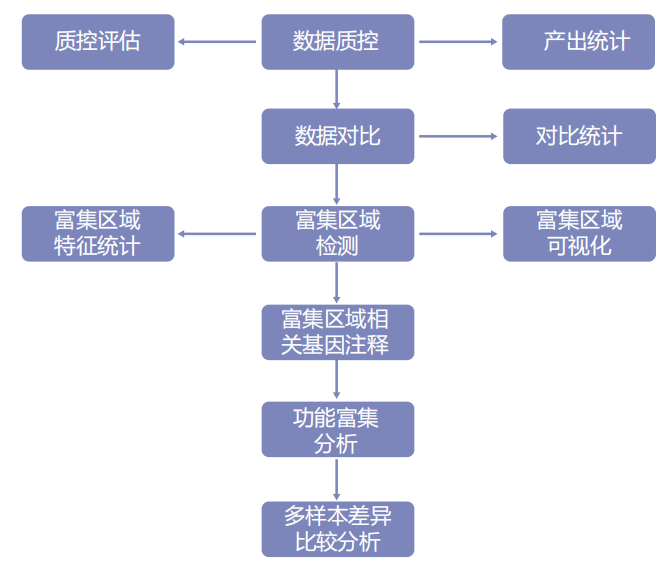
Information analysis
Sample parameters
| Sample requirements | Project cycle |
| Type: DNA sample, cell, or tissue; Total amount: ≥ 10 ng chip 7 ed DNA (human samples can be as low as 5ng) or ≥ 10 cell amounts; DNA fragment size: distributed in the range of 100-500 BP, and the main band is obvious. It is necessary to provide the detection gel map after DNA disruption; Chip standard antibodies: h3k4me3, h3k4me2, h3k9me3, H3K9me2, H3K27me3, h3k27me2, etc. recommended data volume: 20m~40m clean reads. | For DNA samples with less than 25 immunoprecipitated samples, the operation cycle of the standard process (including only standard information analysis) is about 30 working days; For less than 12 cell line samples, the operation cycle of the standard process (including only standard information analysis) is about 50 working days; When the number of samples is more, the project cycle depends on the project scale. |
Case analysis
Case:
Histone modifications are associated with altered antigen-specific T cell responses
XM Yang, et al. (2014)Histone modifications are associated with Delta(9)-tetrahydrocannabinol-mediated alterations in antigen- specific T cell responses. J Biol Chem. 2014 May 19. pij: jbc. M113. 545210.
Background:
Cannabis is a drug, but it can also be used as medicine. Among the ingredients of cannabis, Δ 9 tetrahydrocannabinol also has the effect of anti-inflammatory drugs. Whether the immune regulation of Δ 9 tetrahydrocannabinol is regulated by epigenetic factors has not been studied before.
Methods:
Chip SEQ technology was used to study the changes of histone modifications in antigen-specific T cell responses mediated by Δ 9 tetrahydrocannabinol. There are five kinds of antibodies used here, including H3K4, H3K27, H3K9, H3K36 tri methylation and H3K9Ac.
Conclusion:
The results showed that most of the methylated and acetylated regions of histones were changed by THC (Δ 9 tetrahydrocannabinol). Thc activated part of the genes, while repressing the expression of another part of the genes through histone regulation. Functional clustering analysis of these histone marks showed that these different related genes are related to various cellular functions, ranging from cell cycle regulation to metabolism. Thc regulates immune responses by regulating histone modifications.
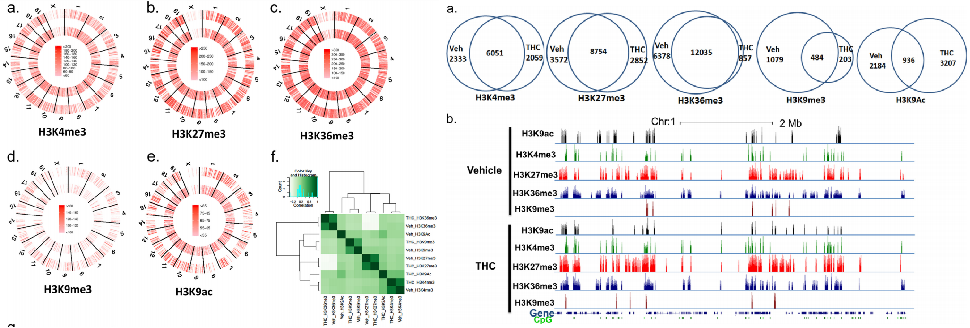
Distribution of H3 histones in lymphoid node cells activates the distribution of H3 histone methylation regions in lymphoid node cells



