Introduction
On the basis of continuing the advantages of 450k single nucleotide resolution detection, Illumina launched an upgraded human genome-wide methylation chip Infinium ® MethylationEPIC BeadChip。 The chip covers more than 850000 CPGs regions and spans all human methylomes. It can detect methylation of many types of samples, including FFPE samples, and truly achieves high-throughput and low-cost. Infinium ® Methylationepic beadchip, with its more comprehensive coverage and high-throughput sample detection ability, has become an ideal research tool for the new season of epigenome Association Research (ewas).
Technical advantages
1. genome wide coverage (>850000 methylation sites)
CpG island non CpG island
Fantom5 enhancer
Encode open chromatin and transcription factor binding site (enhancer region) miRNA promoter region
2. good technical repeatability
98% technical repeatability
The agreement with 450k results was excellent (r> 0.98)
Covers 90% of CpG sites on 450k chips
3. the first methylation chip that can detect FFPE samples
4. High accuracy
The reproducibility of target region enrichment, sequencing depth distribution and coverage is high; Agilent sureselectxt human methyl SEQ capture probe is preferred, and the target regions include:
① The target region was 84m, and the CPG site of 3.7m was detected;
② It mainly targets CpG island, gencode promoter and known regulatory regions;
③ Covering known cancer specific DMR regions and tissue-specific DMR regions;
④ It contains the "shores" and "shells" regions 4KB upstream and downstream of CpG island;
⑤ Contains deoxyribonuclease I (DNase I) high-sensitivity sites. The whole genome coding gene promoter chip (2k upstream and 500bp downstream of TSS, nimblegene chip) can also be used.
Experimental strategy
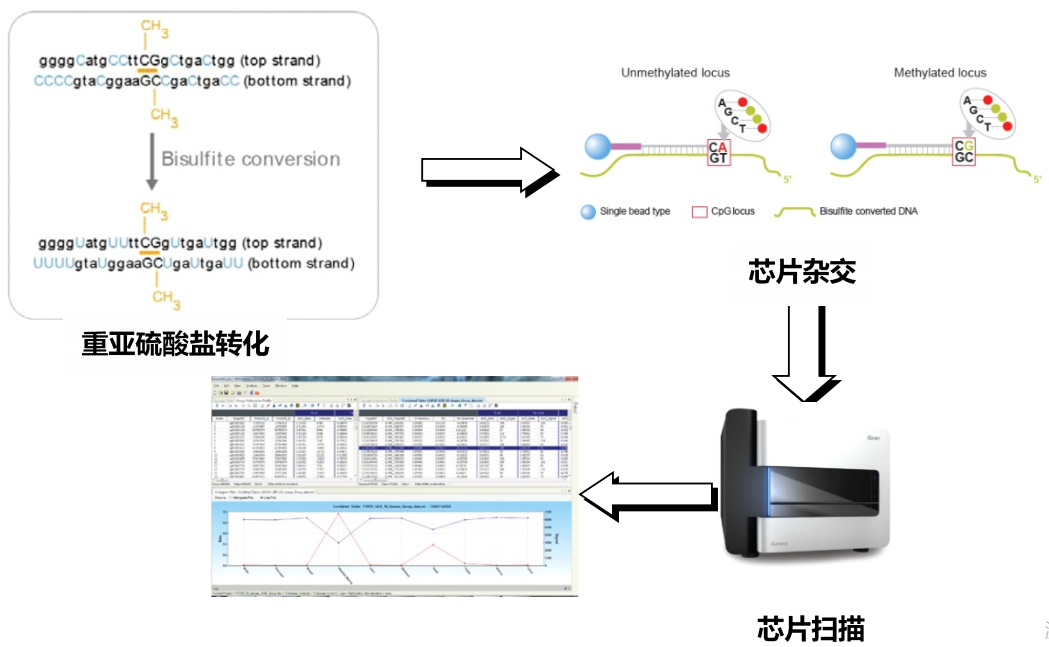
Technical parameter
| Sample requirements | Project cycle |
| 1. sample purity: OD 260/280 value should be between 1.8 and 2.0; RNA should be removed clean; There shall be no DNA contamination from other individuals or other species. two Sample concentration: the concentration shall not be lower than 55ng/ μ L. three Total amount of samples: the total amount of each sample shall not be less than 2 μ G. | 40 working days |
Case analysis
Case: whole genome methylation sequencing analysis of the relationship between age and DNA methylation
Heyn H, Li N, Ferreira HJ, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA. 2012 Jun 26;109(26):10522-7.)
Background:
Human life span is not only determined by genetics, environmental factors can have a non negligible impact on the process of aging by changing epigenetic modifications. The DNA samples of a 103 year old man and a newborn boy were compared by wgbs technology, and the methylation chip was verified in a larger sample size.
Methods:
RNA and DNA were extracted from peripheral blood of healthy elderly (Y103) and umbilical cord blood of newborn (NB), respectively. RNA and DNA were extracted from blood, respectively. DNA was interrupted and bisulfite processed to build a database, then sequenced, and finally verified with a large sample size methylation chip.
Conclusion:
It was finally found that there were more unmethylated sites in centenarians' DNA as a whole compared with newborns of the same cell type.
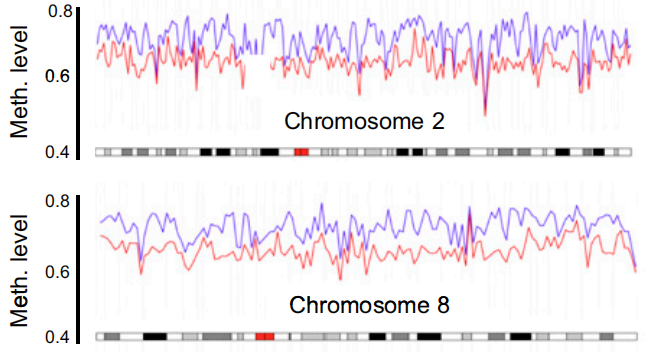
Methylation distribution of centenarians (red) and newborns (blue)



