Introduction
Whole genome bisulfite sequencing (wgbs) is regarded as the "gold standard" of DNA methylation research. Combined with bisulfite treatment and high-throughput sequencing technology, it can realize the methylation analysis of a single c base in the whole genome, and is suitable for the construction of genome-wide fine methylation map.
Conventional whole genome methylation sequencing technology uses t4-dna ligase to connect the linker sequence at both ends of the genomic DNA fragment by ultrasound. The ligation product converts unmethylated cytosine C to uracil u by bisulfite treatment, and then converts uracil u to thymine t by PCR technology mediated by the linker sequence.
Technical advantages
Wide application range
Suitable for human and most animal and plant studies (reference genome)
Genome wide coverage
Maximize access to complete genome-wide methylation information and accurately map methylation
Single base resolution
It can accurately analyze the methylation status of each C base
Experimental strategy

Information analysis
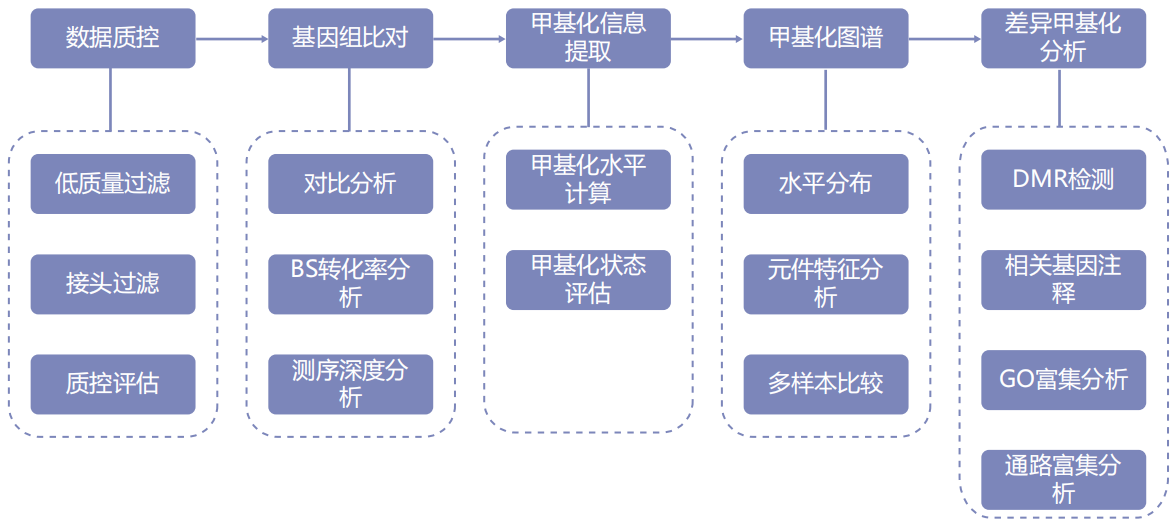
Technical parameter
| Sample requirements | Project cycle |
| Sample DNA requirements: different samples have different requirements, human samples ≥ 2 μ g; Sample DNA Purity: od260/280 = 1.8~2.0; Sample DNA integrity: there is no obvious degradation of DNA, and gel electrophoresis detection gel map shall be provided; Other precautions: ① qubit is used to detect DNA samples, and the final amount of DNA is subject to our detection results; ② If the total amount of DNA exceeds the sample requirements, the sample can be required to be preserved; After the project is completed, the remaining samples will not be saved. | The standard operation cycle below 15 samples is about 60 working days. Because different species require different amounts of data, and the number of databases required varies greatly, the project cycle needs to be determined with the technical support personnel in case of a large number of samples. |
Case analysis
Case: methylomics study of mouse embryonic stem cells in two states
10
Whole-Genome Bisulfile Sequencing of Two Distinct Interconvertible DNA Methylomes of Mouse Embryonic Stem Cells. Cell Stem Cell(2013),13(3),360-369.
Background:
Mouse embryonic stem cells generally grow in a matrix containing serum, known as serum stem cells (serum ESCs); The addition of two kinase inhibitors enables embryonic stem cells to maintain the ground state of pluripotency in the absence of serum. Such stem cells are called 2I stem cells (2iescs); These two states of embryonic stem cells can be transformed into each other. Previous methylation studies in this area were mostly based on mass spectrometry, with limited coverage and research results, and there was still a lack of methylomics studies of 2I embryonic stem cells.
Methods:
利用全基因组Bisulfite甲基化测序,对这两种可互相转换的小鼠胚胎干细胞进行甲基化组学研究。
Conclusion:
The DNA methylation modification of two kinds of mouse embryonic stem cells was comprehensively and accurately detected and systematically compared; Compared with serum ESCs, male 2iescs were globally hypomethylated; In serum, female ESCs showed global hypomethylation similar to male 2iescs, while in the state of 2iescs, the methylation level would further decrease.
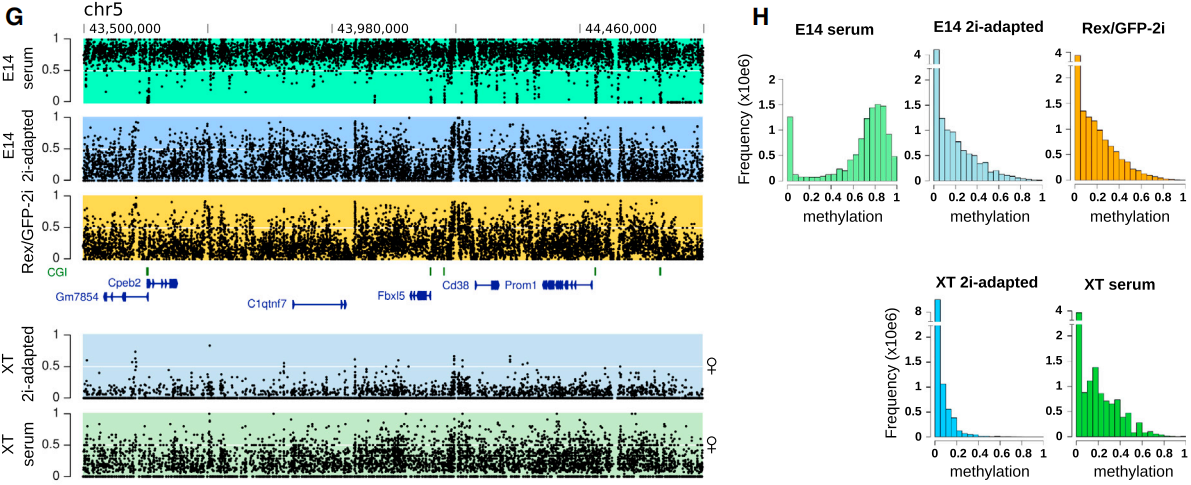
Comparison of methylation modification of mouse embryonic stem cells in different states


