Introduction
Reduced representation bisulfite sequencing (RRBs) uses restriction enzymes to digest the genome, enrich promoters, CpG islands and other important epigenetic regulatory regions, and perform bisulfite sequencing. This technology significantly improves the sequencing depth of high CpG regions, and can obtain high-precision resolution in CpG islands, promoter regions and enhancer element regions. It is an accurate, efficient and economical method for DNA methylation research; It has a broad application prospect in the study of large-scale clinical samples. In order to meet the needs of scientific research technology, we further developed a dual enzyme digestion RRBs (drrbs) that can capture CpG sites in a larger region, which can study the methylation of a wider region, including CGI shore and other regions.
Technical advantages
1. high accuracy
Single base resolution within its coverage
2. good repeatability
The reproducibility of the coverage area of the diversity book can reach 85%-95%, which is suitable for the difference analysis among multiple samples
3. high cost performance
Sequencing regions targeted high CpG regions, resulting in higher data utilization
Experimental strategy
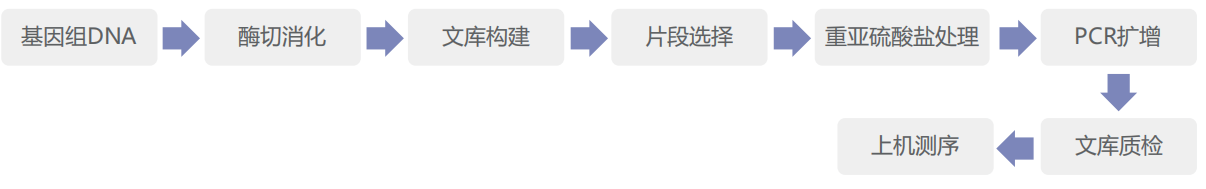
Information analysis
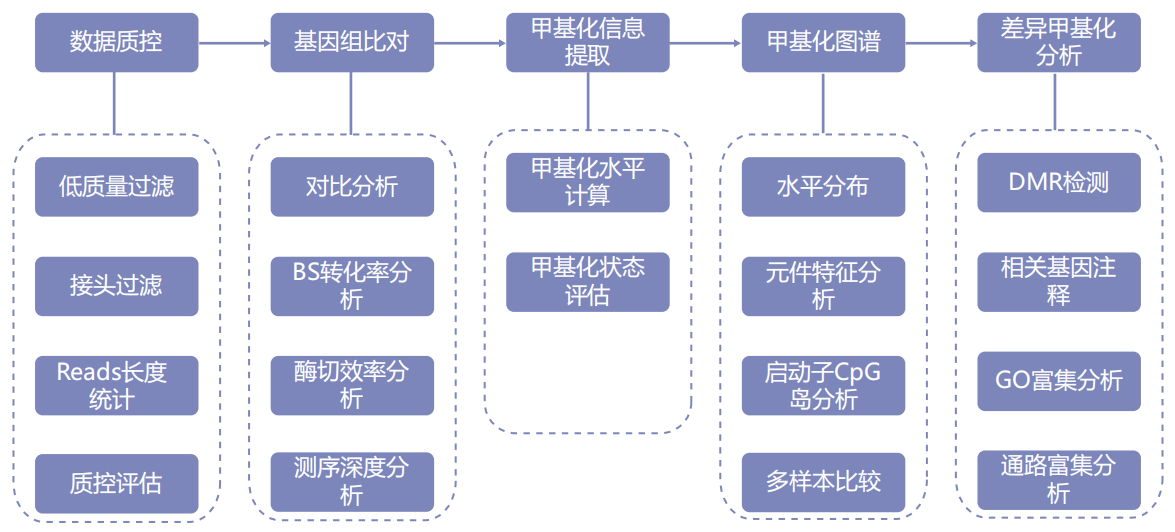
Technical parameter
| Sample requirements | Project cycle |
| Total amount: different samples have different requirements, human samples ≥ 2 μ g; Purity: od260/280 = 1.8~2.0; Integrity: there is no obvious degradation of DNA, and gel electrophoresis detection gel map shall be provided. Recommended data volume: 10g clean data | The operation cycle of the standard process below 15 samples is about 60 working days. When the number of samples is large, the project cycle needs to be determined with the technical support personnel. |
Case analysis
Case:
Effect of DNA methylation changes on the invasive ability of cancer cells
Antje Hascher, et al. (2013) DNA Methyltrans-ferase Inhibition Reverses Epigenetically Embedded Phenotypes in Lung Cancer Preferentially Affecting Polycomb Target Genes. Clin Cancer Res; 20(4); 814–26.
Background:
The phenotype of cancer cells is determined to some extent by the appearance, such as DNA methylation. The metastatic properties in the later stage of cancer development may be related to the changes of epigenetic modifications.
Methods:
RRBs technology was used to detect the methylation modification of highly invasive non-small cell lung cancer cell lines (A549 and htb56) and the corresponding cell lines treated with methylation inhibitor azacytidine. Objective to investigate the effect of methylation changes on the invasive ability of lung cancer cell lines.
Conclusion:
During the development of highly invasive cell lines, DNA methylation modification has undergone extensive changes. Compared with low invasive cell lines, 2.5% of CpG enriched regions detected by RRBs were differentially modified. When azacytidine, a DNA methylation inhibitor, was used, with the loss of methylation modification at these hypermethylated sites, the invasive ability of cell lines was also reversed.
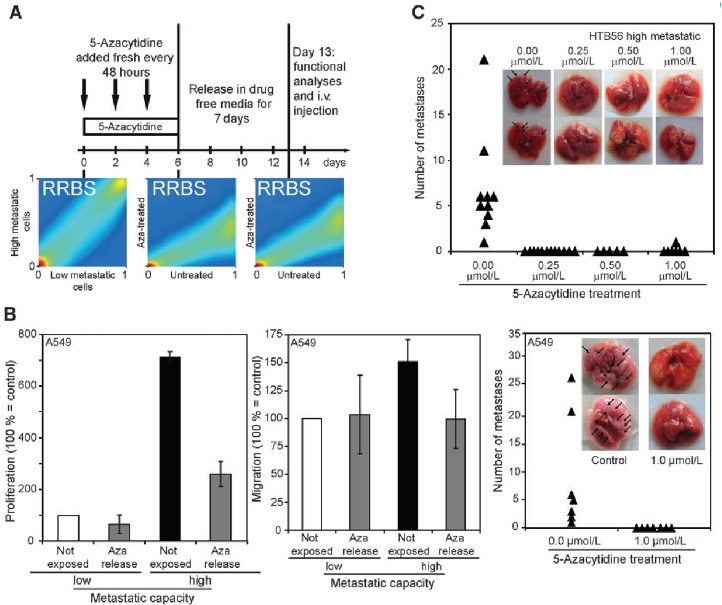
Reversal of invasive ability induced by 5-azacytidine



