Introduction
Methylated DNA immunoprecipitation sequencing (medip SEQ) uses 5 '- methylcytosine specific antibody to enrich methylated DNA fragments, and then combines with high-throughput sequencing technology to quickly and efficiently search for methylated regions at the whole genome level with a small amount of data, so as to compare DNA methylation differences between different cells, tissues and other samples. It can be widely used in disease research and molecular breeding research with large sample size.
Technical advantages
1. wide detection range
Genome wide identification of methylation modified regions
2. targeted
Targeted detection of regions with methylation modifications in the genome
3. low cost
The amount of sequencing data is greatly reduced and the sequencing cost is low
Experimental strategy

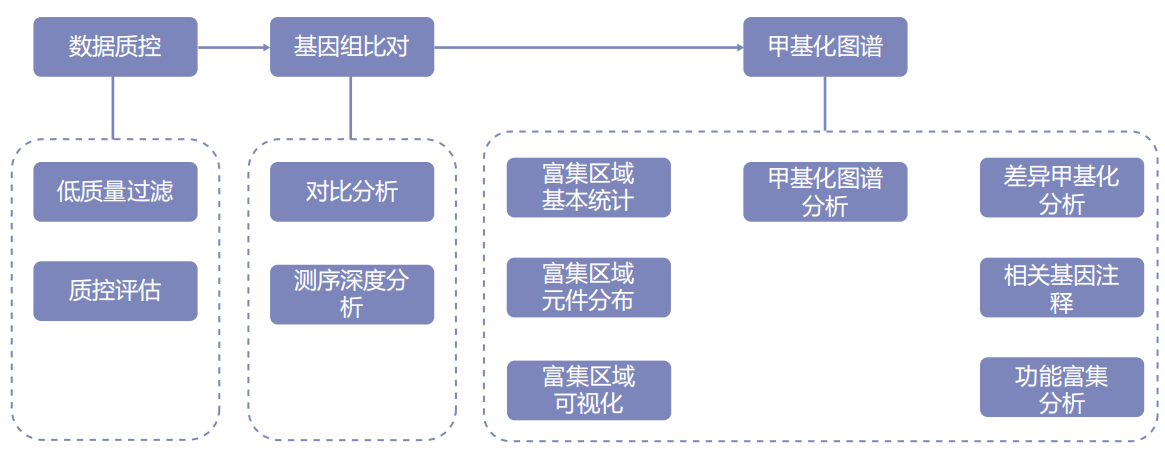
Information analysis
Technical parameter
| Sample requirements | Project cycle |
| Total amount: different samples have different requirements, human samples ≥ 2 μ g; Purity: od260/280 = 1.8~2.0; Integrity: there is no obvious degradation of DNA, and gel electrophoresis detection gel map shall be provided. Recommended data volume: 4G clean data | The operation cycle of the standard process below 15 samples is about 30 working days. When the number of samples is large, the project cycle needs to be determined with the technical support personnel. |
Case analysis
Case: Methylome of porcine fat and muscle tissue
Li, M. et al.An atlas of DNA methylomes in porcine adipose and muscle tissues. Nat. Commun. 3:850 doi: 10.1038/ncomms 1854 (2012).
Background:
Studies have shown that epigenetic modifiers, especially DNA methylation, play an important role in the process of obesity. As an important animal model, pigs are helpful to study human metabolism and obesity.
Methods:
A total of 180 samples from 8 adipose tissues and 2 muscle tissues of 3 kinds of pigs were detected for methylation modification by medip SEQ sequencing technology. Systematically study the relationship between DNA methylation and obesity.
Conclusion:
The DNA methylation profiles of porcine fat and muscle were constructed, and the differentially methylated modified regions (DMRs) among species, sex and tissues were identified. The DMRs in the promoter region were highly correlated with the occurrence of obesity by inhibiting the expression of genes.
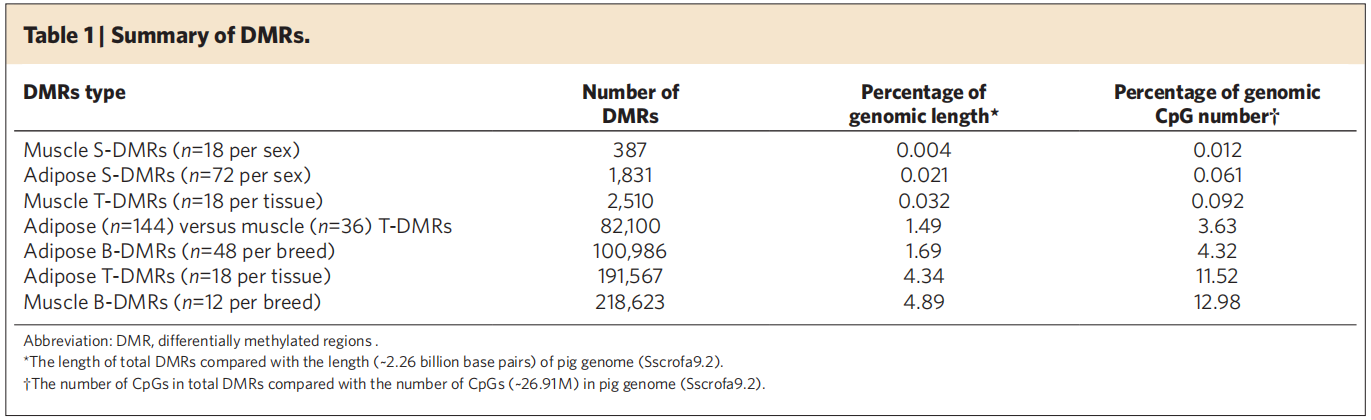
Identified differentially methylated modified regions (DMRs)


